DAD Data Processing: Unterschied zwischen den Versionen
Cbier (Diskussion | Beiträge) Keine Bearbeitungszusammenfassung |
Cbier (Diskussion | Beiträge) Keine Bearbeitungszusammenfassung |
||
| Zeile 1: | Zeile 1: | ||
Displays the DAD Data Processing dialog only when a DAD detector is selected for the Method Configuration of the current Method. | Displays the DAD Data Processing dialog only when a DAD detector is selected for the Method Configuration of the current Method. | ||
Tool Bar Shortcut | '''Tool Bar Shortcut''' | ||
Click on [[File:Data_Processing.png]] . | Click on [[File:Data_Processing.png]] . | ||
{| class="wikitable" | |||
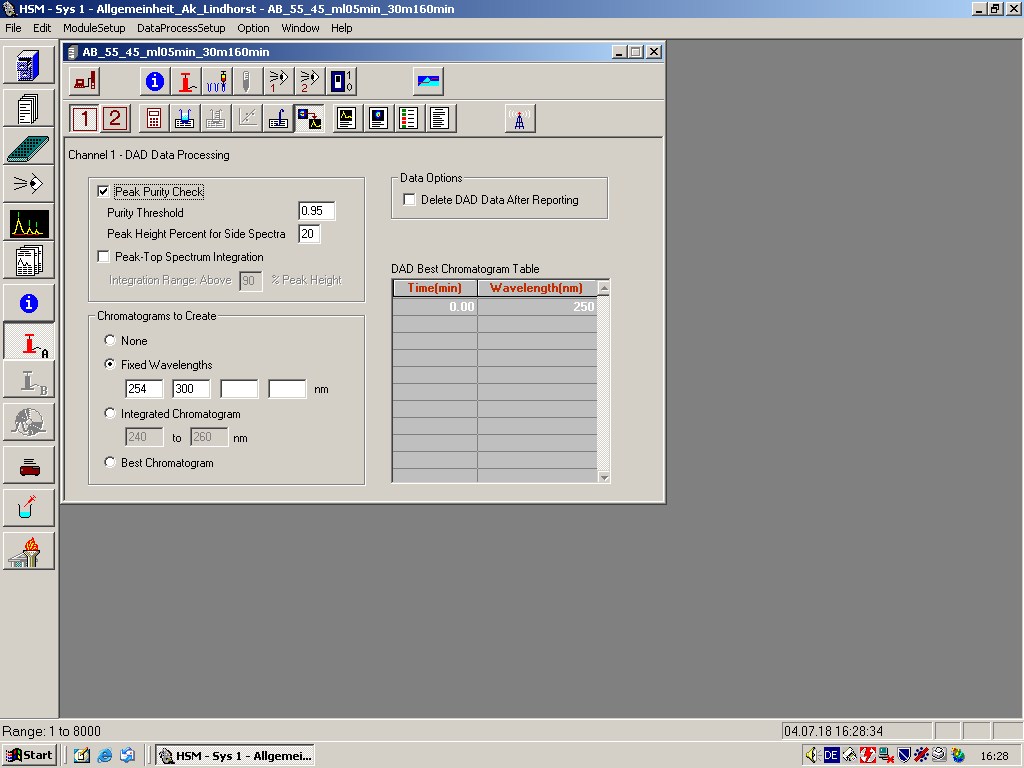
Peak Purity Check If this box is checked, the HSM does a Peak Purity Check. Purity Threshold: Select a value from 0.5 to 1.0. The default is 0.95. A peak with a purity value greater than the specified value is judged to be pure. See Peak Purity Check Using Spectra.Peak Height Percentage for Side Spectra: Specify the percentage of peak height at which side spectra are obtained. | !'''Dialog Entries''' !! | ||
Peak-Top Spectrum Integration: | |- | ||
Chromatograms to Create None: Select if no chromatogram is to be created.Fixed Wavelengths: Select up to four wavelengths for chromatograms to be extracted from DAD. Enter integer only.Integrated Chromatogram: Specify a wavelength range (integers) for integration. The chromatogram is created by performing an integration of spectra over the specified wavelength range.Best Chromatogram: Allows you to extract a chromatogram at various wavelengths varying with the RT. You must specify the wavelengths in the Best Chromatogram Table. DAD Best Chromatogram Table can be edited only when the Best Chromatogram is selected. Enter Time(min) and Wavelength(nm) at which a section of chromatogram is to be extracted. | |style="width:130px;text-align:left; vertical-align:top;"|'''Peak Purity Check''' If this box is checked, the HSM does a Peak Purity Check. | ||
Data Options Check Delete DAD Data After Reporting if you want to delete DAD data after chromatogram extraction and quantification during on-line processing. If Chromatograms to Create is None, this option is disabled.Note that the HSM ignores this option during recalculation, meaning the HSM does not delete DAD data in postprocessing. You can use Delete function in the Open File dialog. | |style="width:525px;text-align:left; vertical-align:top;"|'''Purity Threshold:''' Select a value from 0.5 to 1.0. The default is 0.95. A peak with a purity value greater than the specified value is judged to be pure. See Peak Purity Check Using Spectra. | ||
'''Peak Height Percentage for Side Spectra:''' Specify the percentage of peak height at which side spectra are obtained. | |||
|- | |||
|style="width:130px;text-align:left; vertical-align:top;"|'''Peak-Top Spectrum Integration:''' | |||
|style="width:525px;text-align:left; vertical-align:top;"| | |||
Specify the percentage of peak height above which the integrated peak-top-spectrum is obtained for each chromatogram peak. | |||
|- | |||
|style="width:130px;text-align:left; vertical-align:top;"|'''Chromatograms to Create ''' | |||
|style="width:525px;text-align:left; vertical-align:top;"| | |||
'''None:''' Select if no chromatogram is to be created. | |||
'''Fixed Wavelengths:''' Select up to four wavelengths for chromatograms to be extracted from DAD. Enter integer only. | |||
'''Integrated Chromatogram:''' Specify a wavelength range (integers) for integration. The chromatogram is created by performing an integration of spectra over the specified wavelength range. | |||
'''Best Chromatogram:''' Allows you to extract a chromatogram at various wavelengths varying with the RT. You must specify the wavelengths in the Best Chromatogram Table. | |||
'''DAD Best Chromatogram Table''' can be edited only when the '''Best Chromatogram''' is selected. Enter '''Time(min)''' and '''Wavelength(nm)''' at which a section of chromatogram is to be extracted. | |||
|- | |||
|style="width:130px;text-align:left; vertical-align:top;"|'''Data Options''' | |||
|style="width:525px;text-align:left; vertical-align:top;"| | |||
Check '''Delete DAD Data After Reporting''' if you want to delete DAD data after chromatogram extraction and quantification during on-line processing. If Chromatograms to Create is None, this option is disabled.Note that the HSM ignores this option during recalculation, meaning the HSM does not delete DAD data in postprocessing. You can use Delete function in the Open File dialog. | |||
|- | |||
|} | |||
[[File:L27.png]] | [[File:L27.png]] | ||
Aktuelle Version vom 9. August 2018, 12:28 Uhr
Displays the DAD Data Processing dialog only when a DAD detector is selected for the Method Configuration of the current Method.
Tool Bar Shortcut
| Dialog Entries | |
|---|---|
| Peak Purity Check If this box is checked, the HSM does a Peak Purity Check. | Purity Threshold: Select a value from 0.5 to 1.0. The default is 0.95. A peak with a purity value greater than the specified value is judged to be pure. See Peak Purity Check Using Spectra.
Peak Height Percentage for Side Spectra: Specify the percentage of peak height at which side spectra are obtained. |
| Peak-Top Spectrum Integration: |
Specify the percentage of peak height above which the integrated peak-top-spectrum is obtained for each chromatogram peak. |
| Chromatograms to Create |
None: Select if no chromatogram is to be created. Fixed Wavelengths: Select up to four wavelengths for chromatograms to be extracted from DAD. Enter integer only. Integrated Chromatogram: Specify a wavelength range (integers) for integration. The chromatogram is created by performing an integration of spectra over the specified wavelength range. Best Chromatogram: Allows you to extract a chromatogram at various wavelengths varying with the RT. You must specify the wavelengths in the Best Chromatogram Table. DAD Best Chromatogram Table can be edited only when the Best Chromatogram is selected. Enter Time(min) and Wavelength(nm) at which a section of chromatogram is to be extracted. |
| Data Options |
Check Delete DAD Data After Reporting if you want to delete DAD data after chromatogram extraction and quantification during on-line processing. If Chromatograms to Create is None, this option is disabled.Note that the HSM ignores this option during recalculation, meaning the HSM does not delete DAD data in postprocessing. You can use Delete function in the Open File dialog. |