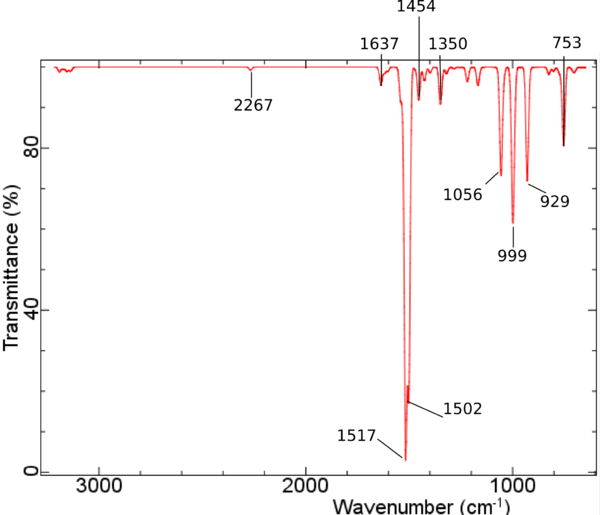
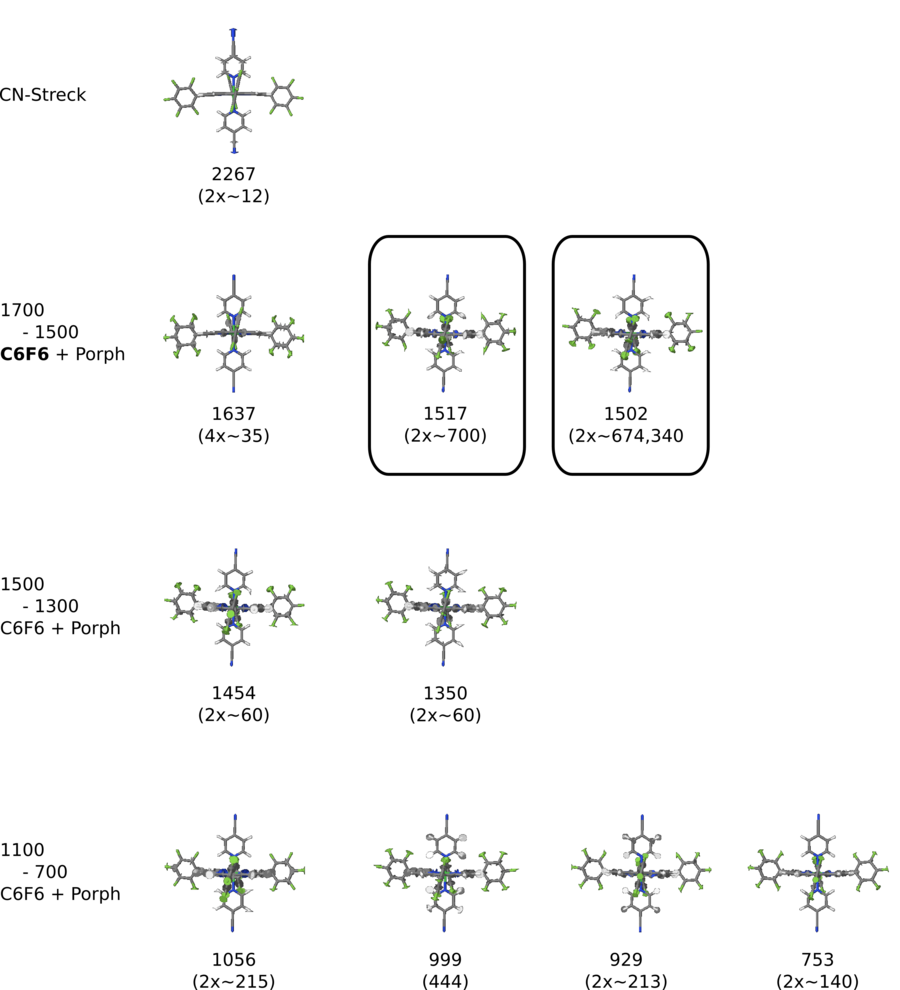
Dicyanopyridin • NiPorph Komplex - IR unskaliert PBE/DZP
zurück: CyanoPorph • Ligand IR-Spektren
L = CyanoPy
- Cyanopyridin IR unskaliert PBE/DZP
- Cyanopyridin IR skaliert PBE/DZP + Experiment
- Cyanopyridin IR experimentell
- Dicyanopyridin • NiPorph Komplex - IR unskaliert PBE/DZP
- Dicyanopyridin • NiPorph Komplex - IR skaliert PBE/DZP + Experiment
- Dicyanopyridin • NiPorph Komplex - IR experimentell
gerechnetes Spektrum (PBE/DZP) des [NiPorph · 2 Cyanopyridin], unskaliert:
Datei:PBE-DZP Niporph Cyanopyridin 2zu1 coplanar-t.IR.pseudo.log
rechte Maustaste/Modell ... zur Auswahl der anzuzeigenden Schwingung
<jmol>
<jmolApplet> <script>
- Jmol state version 11.8.7 2009-08-11 23:55;
function _setWindowState() {
- height 311;
- width 348;
stateVersion = 1108007; backgroundColor = "[x000000]"; axis1Color = "[xff0000]"; axis2Color = "[x008000]"; axis3Color = "[x0000ff]"; ambientPercent = 45; diffusePercent = 84; specular = true; specularPercent = 22; specularPower = 40; specularExponent = 6; statusReporting = true;
}
function _setFileState() {
set allowEmbeddedScripts false;
set autoBond true;
set appendNew true;
set appletProxy "";
set applySymmetryToBonds false;
set bondRadiusMilliAngstroms 150;
set bondTolerance 0.45;
set defaultLattice {0.0 0.0 0.0};
set defaultLoadScript "";
set defaultVDW Jmol;
set forceAutoBond false;
set loadFormat "http://www.rcsb.org/pdb/files/%FILE.pdb";
set minBondDistance 0.4;
set pdbSequential false;
set pdbGetHeader false;
set percentVdwAtom 20;
set smartAromatic true;
# load "http://134.245.135.211/mediawiki/index.php5/Datei:PBE-DZP_Niporph_Cyanopyridin_2zu1_coplanar-t.IR.pseudo.log";
}
function _setVariableState() {
set defaultanglelabel "%VALUE %UNITS";
set defaultcolorscheme "Jmol";
set defaultdistancelabel "%VALUE %UNITS";
set defaultdrawarrowscale 0.5;
set defaultlattice "{0 0 0}";
set defaultloadscript "";
set defaulttorsionlabel "%VALUE %UNITS";
set defaulttranslucent 0.5;
set defaultvdw "Jmol";
set allowembeddedscripts true;
set allowkeystrokes false;
set allowrotateselected false;
set appletproxy "";
set applysymmetrytobonds false;
set atompicking true;
set atomtypes "";
set autobond true;
set autofps false;
set autoloadorientation false;
set axes window;
set axesmode 0;
set axesscale 2.0;
set bondmodeor false;
set bondpicking false;
set bondradiusmilliangstroms 150;
set bondtolerance 0.45;
set cartoonrockets false;
set chaincasesensitive false;
set dataseparator "~~~";
set delaymaximumms 0;
set dipolescale 1.0;
set disablepopupmenu false;
set displaycellparameters true;
set dotdensity 3;
set dotsselectedonly false;
set dotsurface true;
set dragselected false;
set drawhover false;
set drawpicking false;
set dynamicmeasurements false;
set ellipsoidarcs false;
set ellipsoidaxes false;
set ellipsoidaxisdiameter 0.02;
set ellipsoidball true;
set ellipsoiddotcount 200;
set ellipsoiddots false;
set ellipsoidfill false;
set fontcaching true;
set forceautobond false;
set greyscalerendering false;
set hbondsangleminimum 90.0;
set hbondsbackbone false;
set hbondsdistancemaximum 3.25;
set hbondssolid false;
set helixstep 1;
set helppath "http://chemapps.stolaf.edu/jmol/docs/index.htm";
set hermitelevel 0;
set hidenameinpopup false;
set hidenavigationpoint false;
set highresolution false;
set historylevel 0;
set hoverdelay 0.5;
set imagestate true;
set isosurfacepropertysmoothing true;
set justifymeasurements false;
set loadatomdatatolerance 0.01;
set loadformat "http://www.rcsb.org/pdb/files/%FILE.pdb";
set measureallmodels false;
set measurementlabels true;
set messagestylechime false;
set minbonddistance 0.4;
set navigatesurface false;
set navigationperiodic false;
set navigationspeed 5.0;
set pdbgetheader false;
set pdbsequential false;
set percentvdwatom 20;
set pickingspinrate 10;
set picklabel "";
set pointgroupdistancetolerance 0.2;
set pointgrouplineartolerance 8.0;
set propertyatomnumbercolumncount 0;
set propertyatomnumberfield 0;
set propertycolorscheme "roygb";
set propertydatacolumncount 0;
set propertydatafield 0;
set quaternionframe "p";
set rangeselected false;
set ribbonaspectratio 16;
set ribbonborder false;
set rocketbarrels false;
set selectallmodels true;
set selecthetero true;
set selecthydrogen true;
set sheetsmoothing 1.0;
set showhiddenselectionhalos false;
set showhydrogens true;
set showkeystrokes true;
set showmeasurements true;
set showmultiplebonds true;
set shownavigationpointalways false;
set smartaromatic true;
set solventprobe false;
set solventproberadius 1.2;
set ssbondsbackbone false;
set stereodegrees -5;
set strandcountformeshribbon 7;
set strandcountforstrands 5;
set testflag1 false;
set testflag2 false;
set testflag3 false;
set testflag4 false;
set tracealpha true;
set useminimizationthread true;
set usenumberlocalization true;
set vectorscale 2.0;
set vibrationperiod 0.5;
set vibrationscale 0.5;
set wireframerotation false;
set zoomlarge true;
set zshade false;
- user-defined variables;
- --no global user variables defined--;
- label defaults;
select none; color label none; background label none; set labelOffset 4 4; set labelAlignment left; set labelPointer off; font label 13.0 SansSerif Plain;
}
function _setModelState() {
select ({0:30703});
Spacefill 0.0;
select BONDS ({0:34655});
wireframe 0.12;
measures delete;
select *; set measures nanometers;
font measures 15.0 SansSerif Plain;
select measures ({null});
select ({0:30703});
Vectors 0.0030;
boundBox off;
frank on;
font frank 16.0 SansSerif Bold;
frame 1.1; moveto -1.0 { 864 353 -359 99.45} 100.0 0.0 0.16 {0.0 0.0 0.02210617} 10.838539 {0.0 0.0 0.0} 0.0013241823 -0.11343511 0.0;
frame 1.287; moveto -1.0 { 864 353 -359 99.45} 100.0 0.0 0.16 {0.0 0.0 0.02210617} 10.838539 {0.0 0.0 0.0} 0.0013241823 -0.11343511 0.0;
set fontScaling false;
}
function _setFrameState() {
- frame state;
- modelCount 304;
- first 1.1;
- last 1.304;
frame RANGE 1.1 1.304; animation DIRECTION +1; animation FPS 10; animation MODE ONCE 0.0 0.0; frame 1.288; animation OFF;
}
function _setPerspectiveState() {
set perspectiveModel 11;
set scaleAngstromsPerInch 0.0;
set perspectiveDepth true;
set visualRange 5.0;
set cameraDepth 3.0;
boundbox corners {-6.422249 -6.428316 -7.5961423} {6.422249 6.428316 7.6403546} # volume = 2516.1091;
center {0.0 0.0 0.02210617};
moveto 0.0 { 873 355 -334 101.53} 100.0 0.0 0.16 {0.0 0.0 0.02210617} 10.838539 {0.0 0.0 0.0} 2.4554372E-4 -0.11321433 0.0;;
slab 100;depth 0;
set spinX 0; set spinY 30; set spinZ 0; set spinFps 30; set navX 0; set navY 0; set navZ 0; set navFps 10;
vibration ON;
}
function _setSelectionState() {
select ({0:30703});
set hideNotSelected false;
}
function _setState() {
initialize; set refreshing false; _setWindowState; _setFileState; _setVariableState; _setModelState; _setFrameState; _setPerspectiveState; _setSelectionState; set refreshing true; set antialiasDisplay false; set antialiasTranslucent true; set antialiasImages true;
}
_setState;
</script> <uploadedFileContents>PBE-DZP_Niporph_Cyanopyridin_2zu1_coplanar-t.IR.pseudo.log</uploadedFileContents> </jmolApplet>
</jmol> CNPy-Porph Streckschwingung bei 2267 Wellenzahlen
Datei:Niporph Cyanopyridin 2zu1 coplanar-t IR unskaliert mit banden.zip
Datei:Niporph Cyanopyridin 2zu1 coplanar-t IR schwingungen.txt