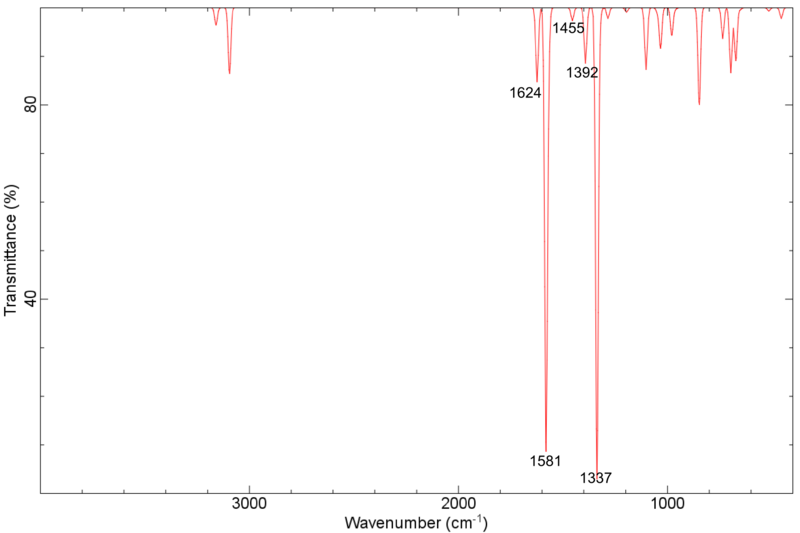
Nitropyridin IR unskaliert PBE/DZP
Zur Navigation springen
Zur Suche springen
zurück: NiPorph • Nitropy IR-Spektren
L = NitroPy
- Nitropyridin IR unskaliert PBE/DZP
- Nitropyridin IR skaliert PBE/DZP + Experiment
- Nitropyridin IR experimentell
- (*obsolet*) - verschiedene Versuche zum Vergleich gerechnetes zu gemessenem Spektrum des Nitropyridin
- Dinitropyridin • NiPorph Komplex - IR unskaliert PBE/DZP
- Dinitropyridin • NiPorph Komplex - IR skaliert PBE/DZP + Experiment
- Dinitropyridin • NiPorph Komplex - IR experimentell
gerechnetes Spektrum (PBE/DZP) des Nitropyridin, unskaliert:
Datei:PBE-DZP Nitropyridin IR.pseudo.log
rechte Maustaste/Modell ... zur Auswahl der anzuzeigenden Schwingung
<jmol>
<jmolApplet> <script>
- Jmol state version 11.8.7 2009-08-11 23:55;
function _setWindowState() {
- height 397;
- width 477;
stateVersion = 1108007; backgroundColor = "[x000000]"; axis1Color = "[xff0000]"; axis2Color = "[x008000]"; axis3Color = "[x0000ff]"; ambientPercent = 45; diffusePercent = 84; specular = true; specularPercent = 22; specularPower = 40; specularExponent = 6; statusReporting = true;
}
function _setFileState() {
set allowEmbeddedScripts false;
set autoBond true;
set appendNew true;
set appletProxy "";
set applySymmetryToBonds false;
set bondRadiusMilliAngstroms 150;
set bondTolerance 0.45;
set defaultLattice {0.0 0.0 0.0};
set defaultLoadScript "";
set defaultVDW Jmol;
set forceAutoBond false;
set loadFormat "http://www.rcsb.org/pdb/files/%FILE.pdb";
set minBondDistance 0.4;
set pdbSequential false;
set pdbGetHeader false;
set percentVdwAtom 20;
set smartAromatic true;
}
function _setVariableState() {
set defaultanglelabel "%VALUE %UNITS";
set defaultcolorscheme "Jmol";
set defaultdistancelabel "%VALUE %UNITS";
set defaultdrawarrowscale 0.5;
set defaultlattice "{0 0 0}";
set defaultloadscript "";
set defaulttorsionlabel "%VALUE %UNITS";
set defaulttranslucent 0.5;
set defaultvdw "Jmol";
set allowembeddedscripts true;
set allowkeystrokes false;
set allowrotateselected false;
set appletproxy "";
set applysymmetrytobonds false;
set atompicking true;
set atomtypes "";
set autobond true;
set autofps false;
set autoloadorientation false;
set axes window;
set axesmode 0;
set axesscale 2.0;
set bondmodeor false;
set bondpicking false;
set bondradiusmilliangstroms 150;
set bondtolerance 0.45;
set cartoonrockets false;
set chaincasesensitive false;
set dataseparator "~~~";
set delaymaximumms 0;
set dipolescale 1.0;
set disablepopupmenu false;
set displaycellparameters true;
set dotdensity 3;
set dotsselectedonly false;
set dotsurface true;
set dragselected false;
set drawhover false;
set drawpicking false;
set dynamicmeasurements false;
set ellipsoidarcs false;
set ellipsoidaxes false;
set ellipsoidaxisdiameter 0.02;
set ellipsoidball true;
set ellipsoiddotcount 200;
set ellipsoiddots false;
set ellipsoidfill false;
set fontcaching true;
set forceautobond false;
set greyscalerendering false;
set hbondsangleminimum 90.0;
set hbondsbackbone false;
set hbondsdistancemaximum 3.25;
set hbondssolid false;
set helixstep 1;
set helppath "http://chemapps.stolaf.edu/jmol/docs/index.htm";
set hermitelevel 0;
set hidenameinpopup false;
set hidenavigationpoint false;
set highresolution false;
set historylevel 0;
set hoverdelay 0.5;
set imagestate true;
set isosurfacepropertysmoothing true;
set justifymeasurements false;
set loadatomdatatolerance 0.01;
set loadformat "http://www.rcsb.org/pdb/files/%FILE.pdb";
set measureallmodels false;
set measurementlabels true;
set messagestylechime false;
set minbonddistance 0.4;
set navigatesurface false;
set navigationperiodic false;
set navigationspeed 5.0;
set pdbgetheader false;
set pdbsequential false;
set percentvdwatom 20;
set pickingspinrate 10;
set picklabel "";
set pointgroupdistancetolerance 0.2;
set pointgrouplineartolerance 8.0;
set propertyatomnumbercolumncount 0;
set propertyatomnumberfield 0;
set propertycolorscheme "roygb";
set propertydatacolumncount 0;
set propertydatafield 0;
set quaternionframe "p";
set rangeselected false;
set ribbonaspectratio 16;
set ribbonborder false;
set rocketbarrels false;
set selectallmodels true;
set selecthetero true;
set selecthydrogen true;
set sheetsmoothing 1.0;
set showhiddenselectionhalos false;
set showhydrogens true;
set showkeystrokes true;
set showmeasurements true;
set showmultiplebonds true;
set shownavigationpointalways false;
set smartaromatic true;
set solventprobe false;
set solventproberadius 1.2;
set ssbondsbackbone false;
set stereodegrees -5;
set strandcountformeshribbon 7;
set strandcountforstrands 5;
set testflag1 false;
set testflag2 false;
set testflag3 false;
set testflag4 false;
set tracealpha true;
set useminimizationthread true;
set usenumberlocalization true;
set vectorscale 3.0;
set vibrationperiod 0.5;
set vibrationscale 0.5;
set wireframerotation false;
set zoomlarge true;
set zshade false;
- user-defined variables;
- --no global user variables defined--;
- label defaults;
select none; color label none; background label none; set labelOffset 4 4; set labelAlignment left; set labelPointer off; font label 13.0 SansSerif Plain;
}
function _setModelState() {
select ({0:519});
Spacefill 0.0;
select BONDS ({0:519});
wireframe 0.04;
measures delete;
select *; set measures nanometers;
font measures 15.0 SansSerif Plain;
select measures ({null});
select ({0:519});
Vectors 0.1;
boundBox off;
frank on;
font frank 16.0 SansSerif Bold;
frame 1.1; moveto -1.0 { -999 -40 -36 93.49} 100.0 0.1 0.13 {0.0 0.0 0.27606106} 4.35575 {0.0 0.0 0.0} 0.0061642947 3.7868779 0.0;
frame 1.2; moveto -1.0 { -999 -40 -36 93.49} 100.0 0.1 0.13 {0.0 0.0 0.27606106} 4.35575 {0.0 0.0 0.0} 0.0061642947 3.7868779 0.0;
set fontScaling false;
}
function _setFrameState() {
- frame state;
- modelCount 40;
- first 1.1;
- last 1.40;
frame RANGE 1.1 1.40; animation DIRECTION +1; animation FPS 10; animation MODE ONCE 0.0 0.0; frame 1.35; animation OFF;
}
function _setPerspectiveState() {
set perspectiveModel 11;
set scaleAngstromsPerInch 0.0;
set perspectiveDepth true;
set visualRange 5.0;
set cameraDepth 3.0;
boundbox corners {-2.1618116 0.0 -2.1425958} {2.1618116 0.0 2.694718} # volume = 0.0;
center {0.0 0.0 0.27606106};
moveto 0.0 { -999 -40 -36 93.49} 100.0 0.1 0.13 {0.0 0.0 0.27606106} 4.35575 {0.0 0.0 0.0} 0.0061642947 3.7868779 0.0;;
slab 100;depth 0;
set spinX 0; set spinY 30; set spinZ 0; set spinFps 30; set navX 0; set navY 0; set navZ 0; set navFps 10;
vibration ON;
}
function _setSelectionState() {
select ({0:519});
set hideNotSelected false;
}
function _setState() {
initialize; set refreshing false; _setWindowState; _setFileState; _setVariableState; _setModelState; _setFrameState; _setPerspectiveState; _setSelectionState; set refreshing true; set antialiasDisplay false; set antialiasTranslucent true; set antialiasImages true;
}
_setState;
</script> <uploadedFileContents>PBE-DZP_Nitropyridin_IR.pseudo.log</uploadedFileContents> </jmolApplet>
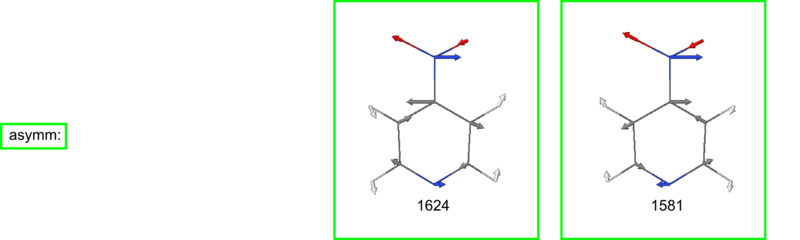
</jmol> Nitropy asymm Streckschwingung des NO2 bei 1581 Wellenzahlen
$vibrational spectrum
# mode symmetry wave number IR intensity selection rules
# cm**(-1) km/mol IR RAMAN
1 0.00 0.00000 - -
2 0.00 0.00000 - -
3 0.00 0.00000 - -
4 0.00 0.00000 - -
5 0.00 0.00000 - -
6 0.00 0.00000 - -
7 a2 66.66 0.00000 NO YES
8 b2 165.73 5.94685 YES YES
9 b1 243.31 2.97145 YES YES
10 a1 374.86 0.14606 YES YES
11 a2 375.67 0.00000 NO YES
12 b2 455.60 4.06959 YES YES
13 b1 515.22 1.26300 YES YES
14 b1 653.02 0.74329 YES YES
15 a1 673.08 20.32900 YES YES
16 b2 696.93 25.00909 YES YES
17 b2 735.66 11.82458 YES YES
18 a1 846.94 25.19720 YES YES
19 b2 849.76 13.37935 YES YES
20 a2 863.35 0.00000 NO YES
21 b2 963.20 0.66012 YES YES
22 a2 972.38 0.00000 NO YES
23 a1 979.71 10.55002 YES YES
24 a1 1032.98 15.54438 YES YES
25 b1 1047.93 1.71872 YES YES
26 a1 1102.68 23.78453 YES YES
27 a1 1196.36 1.67217 YES YES
28 b1 1284.56 4.07138 YES YES
29 b1 1322.34 2.84232 YES YES
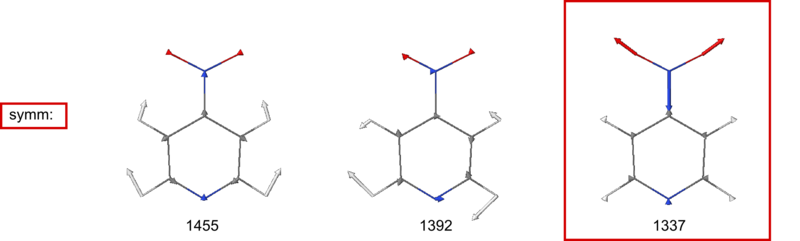
30 a1 1337.33 181.59422 YES YES
31 b1 1392.07 21.35625 YES YES
32 a1 1455.02 4.93500 YES YES
33 a1 1570.00 18.72748 YES YES
34 b1 1580.91 166.83323 YES YES
35 b1 1623.66 28.55283 YES YES
36 b1 3095.07 22.21126 YES YES
37 a1 3097.68 3.43948 YES YES
38 a1 3159.25 4.97279 YES YES
39 b1 3160.47 1.68281 YES YES