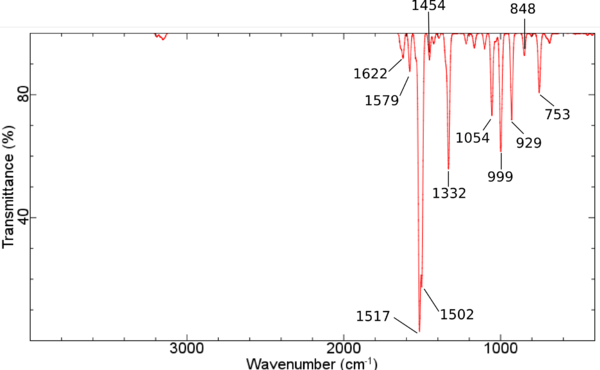
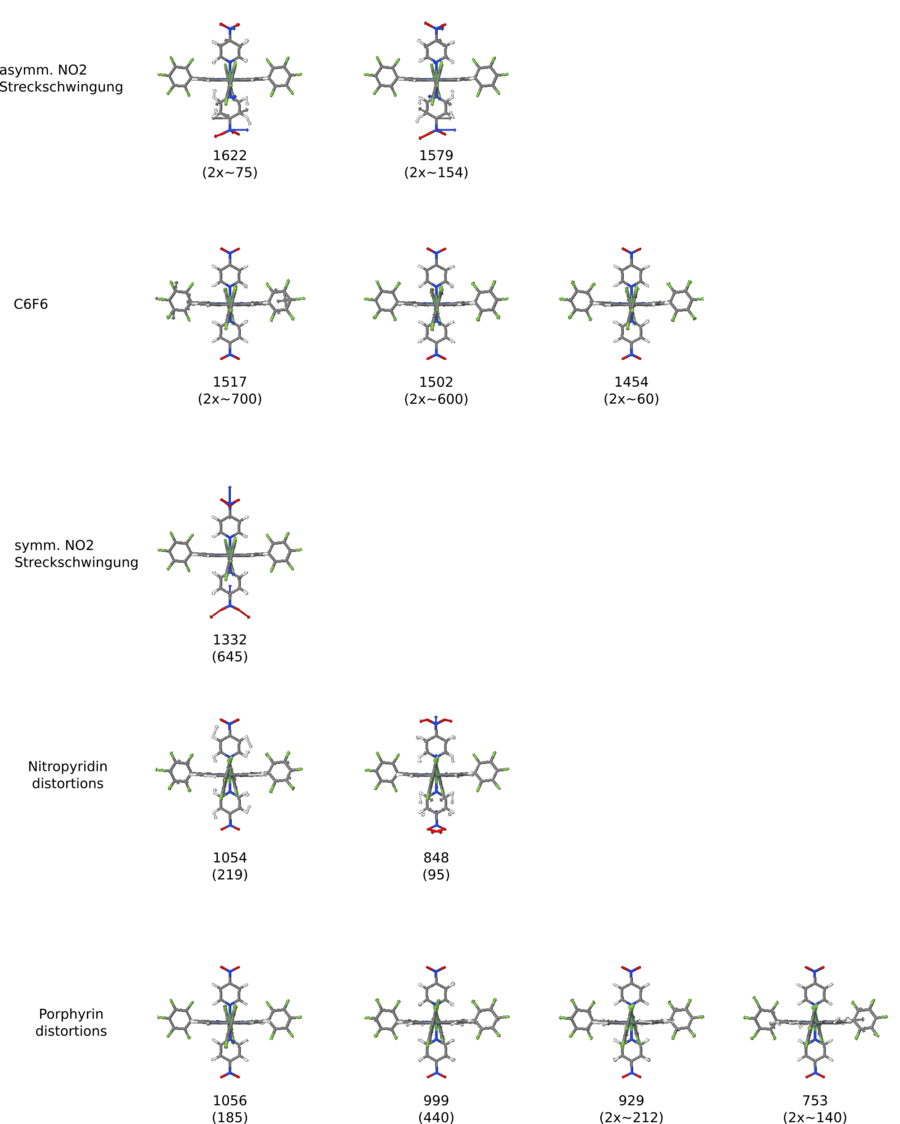
Dinitropyridin • NiPorph Komplex - IR unskaliert PBE/DZP
zurück: NiPorph • Nitropy IR-Spektren
L = NitroPy
- Nitropyridin IR unskaliert PBE/DZP
- Nitropyridin IR skaliert PBE/DZP + Experiment
- Nitropyridin IR experimentell
- (*obsolet*) - verschiedene Versuche zum Vergleich gerechnetes zu gemessenem Spektrum des Nitropyridin
- Dinitropyridin • NiPorph Komplex - IR unskaliert PBE/DZP
- Dinitropyridin • NiPorph Komplex - IR skaliert PBE/DZP + Experiment
- Dinitropyridin • NiPorph Komplex - IR experimentell
gerechnetes Spektrum (PBE/DZP) des [NiPorph · 2 Nitropyridin], unskaliert:
Datei:PBE-DZP Niporph Nitropyridin 2zu1 coplanar-t.IR.pseudo.log
Datei:Niporph Nitropyridin 2zu1 coplanar-t IR unskaliert mit banden.zip
rechte Maustaste/Modell ... zur Auswahl der anzuzeigenden Schwingung
<jmol>
<jmolApplet> <script>
- Jmol state version 11.8.7 2009-08-11 23:55;
function _setWindowState() {
- height 368;
- width 394;
stateVersion = 1108007; backgroundColor = "[x000000]"; axis1Color = "[xff0000]"; axis2Color = "[x008000]"; axis3Color = "[x0000ff]"; ambientPercent = 45; diffusePercent = 84; specular = true; specularPercent = 22; specularPower = 40; specularExponent = 6; statusReporting = true;
}
function _setFileState() {
set allowEmbeddedScripts false;
set autoBond true;
set appendNew true;
set appletProxy "";
set applySymmetryToBonds false;
set bondRadiusMilliAngstroms 150;
set bondTolerance 0.45;
set defaultLattice {0.0 0.0 0.0};
set defaultLoadScript "";
set defaultVDW Jmol;
set forceAutoBond false;
set loadFormat "http://www.rcsb.org/pdb/files/%FILE.pdb";
set minBondDistance 0.4;
set pdbSequential false;
set pdbGetHeader false;
set percentVdwAtom 20;
set smartAromatic true;
}
function _setVariableState() {
set defaultanglelabel "%VALUE %UNITS";
set defaultcolorscheme "Jmol";
set defaultdistancelabel "%VALUE %UNITS";
set defaultdrawarrowscale 0.5;
set defaultlattice "{0 0 0}";
set defaultloadscript "";
set defaulttorsionlabel "%VALUE %UNITS";
set defaulttranslucent 0.5;
set defaultvdw "Jmol";
set allowembeddedscripts true;
set allowkeystrokes false;
set allowrotateselected false;
set appletproxy "";
set applysymmetrytobonds false;
set atompicking true;
set atomtypes "";
set autobond true;
set autofps false;
set autoloadorientation false;
set axes window;
set axesmode 0;
set axesscale 2.0;
set bondmodeor false;
set bondpicking false;
set bondradiusmilliangstroms 150;
set bondtolerance 0.45;
set cartoonrockets false;
set chaincasesensitive false;
set dataseparator "~~~";
set delaymaximumms 0;
set dipolescale 1.0;
set disablepopupmenu false;
set displaycellparameters true;
set dotdensity 3;
set dotsselectedonly false;
set dotsurface true;
set dragselected false;
set drawhover false;
set drawpicking false;
set dynamicmeasurements false;
set ellipsoidarcs false;
set ellipsoidaxes false;
set ellipsoidaxisdiameter 0.02;
set ellipsoidball true;
set ellipsoiddotcount 200;
set ellipsoiddots false;
set ellipsoidfill false;
set fontcaching true;
set forceautobond false;
set greyscalerendering false;
set hbondsangleminimum 90.0;
set hbondsbackbone false;
set hbondsdistancemaximum 3.25;
set hbondssolid false;
set helixstep 1;
set helppath "http://chemapps.stolaf.edu/jmol/docs/index.htm";
set hermitelevel 0;
set hidenameinpopup false;
set hidenavigationpoint false;
set highresolution false;
set historylevel 0;
set hoverdelay 0.5;
set imagestate true;
set isosurfacepropertysmoothing true;
set justifymeasurements false;
set loadatomdatatolerance 0.01;
set loadformat "http://www.rcsb.org/pdb/files/%FILE.pdb";
set measureallmodels false;
set measurementlabels true;
set messagestylechime false;
set minbonddistance 0.4;
set navigatesurface false;
set navigationperiodic false;
set navigationspeed 5.0;
set pdbgetheader false;
set pdbsequential false;
set percentvdwatom 20;
set pickingspinrate 10;
set picklabel "";
set pointgroupdistancetolerance 0.2;
set pointgrouplineartolerance 8.0;
set propertyatomnumbercolumncount 0;
set propertyatomnumberfield 0;
set propertycolorscheme "roygb";
set propertydatacolumncount 0;
set propertydatafield 0;
set quaternionframe "p";
set rangeselected false;
set ribbonaspectratio 16;
set ribbonborder false;
set rocketbarrels false;
set selectallmodels true;
set selecthetero true;
set selecthydrogen true;
set sheetsmoothing 1.0;
set showhiddenselectionhalos false;
set showhydrogens true;
set showkeystrokes true;
set showmeasurements true;
set showmultiplebonds true;
set shownavigationpointalways false;
set smartaromatic true;
set solventprobe false;
set solventproberadius 1.2;
set ssbondsbackbone false;
set stereodegrees -5;
set strandcountformeshribbon 7;
set strandcountforstrands 5;
set testflag1 false;
set testflag2 false;
set testflag3 false;
set testflag4 false;
set tracealpha true;
set useminimizationthread true;
set usenumberlocalization true;
set vectorscale 5.0;
set vibrationperiod 0.5;
set vibrationscale 0.5;
set wireframerotation false;
set zoomlarge true;
set zshade false;
- user-defined variables;
- --no global user variables defined--;
- label defaults;
select none; color label none; background label none; set labelOffset 4 4; set labelAlignment left; set labelPointer off; font label 13.0 SansSerif Plain;
}
function _setModelState() {
select ({0:31929});
Spacefill 0.0;
select BONDS ({0:35959});
wireframe 0.16;
measures delete;
select *; set measures nanometers;
font measures 15.0 SansSerif Plain;
select measures ({null});
select ({0:31929});
Vectors 0.0040;
boundBox off;
frank on;
font frank 16.0 SansSerif Bold;
frame 1.1; moveto -1.0 { -1000 29 12 87.14} 100.0 0.0 0.0 {0.0 0.0 0.020375013} 10.837841 {0.0 0.0 0.0} -0.0016691721 0.09998653 0.0;
frame 1.2; moveto -1.0 { -1000 29 12 87.14} 100.0 0.0 0.0 {0.0 0.0 0.020375013} 10.837841 {0.0 0.0 0.0} -0.0016691721 0.09998653 0.0;
frame 1.270; moveto -1.0 { -1000 29 12 87.14} 100.0 0.0 0.0 {0.0 0.0 0.020375013} 10.837841 {0.0 0.0 0.0} -0.0016691721 0.09998653 0.0;
frame 1.271; moveto -1.0 { -1000 29 12 87.14} 100.0 0.0 0.0 {0.0 0.0 0.020375013} 10.837841 {0.0 0.0 0.0} -0.0016691721 0.09998653 0.0;
frame 1.272; moveto -1.0 { -1000 29 12 87.14} 100.0 0.0 0.0 {0.0 0.0 0.020375013} 10.837841 {0.0 0.0 0.0} -0.0016691721 0.09998653 0.0;
frame 1.273; moveto -1.0 { -1000 29 12 87.14} 100.0 0.0 0.0 {0.0 0.0 0.020375013} 10.837841 {0.0 0.0 0.0} -0.0016691721 0.09998653 0.0;
frame 1.274; moveto -1.0 { -1000 29 12 87.14} 100.0 0.0 0.0 {0.0 0.0 0.020375013} 10.837841 {0.0 0.0 0.0} -0.0016691721 0.09998653 0.0;
frame 1.275; moveto -1.0 { -1000 29 12 87.14} 100.0 0.0 0.0 {0.0 0.0 0.020375013} 10.837841 {0.0 0.0 0.0} -0.0016691721 0.09998653 0.0;
frame 1.276; moveto -1.0 { -1000 29 12 87.14} 100.0 0.0 0.0 {0.0 0.0 0.020375013} 10.837841 {0.0 0.0 0.0} -0.0016691721 0.09998653 0.0;
frame 1.277; moveto -1.0 { -1000 29 12 87.14} 100.0 0.0 0.0 {0.0 0.0 0.020375013} 10.837841 {0.0 0.0 0.0} -0.0016691721 0.09998653 0.0;
frame 1.278; moveto -1.0 { -1000 29 12 87.14} 100.0 0.0 0.0 {0.0 0.0 0.020375013} 10.837841 {0.0 0.0 0.0} -0.0016691721 0.09998653 0.0;
frame 1.279; moveto -1.0 { -1000 29 12 87.14} 100.0 0.0 0.0 {0.0 0.0 0.020375013} 10.837841 {0.0 0.0 0.0} -0.0016691721 0.09998653 0.0;
frame 1.280; moveto -1.0 { -1000 29 12 87.14} 100.0 0.0 0.0 {0.0 0.0 0.020375013} 10.837841 {0.0 0.0 0.0} -0.0016691721 0.09998653 0.0;
frame 1.281; moveto -1.0 { -277 673 685 148.94} 100.0 0.0 0.0 {0.0 0.0 0.020375013} 10.837841 {0.0 0.0 0.0} 4.531163E-4 0.10011922 0.0;
frame 1.282; moveto -1.0 { -277 673 685 148.94} 100.0 0.0 0.0 {0.0 0.0 0.020375013} 10.837841 {0.0 0.0 0.0} 4.531163E-4 0.10011922 0.0;
frame 1.283; moveto -1.0 { -1000 29 12 87.14} 100.0 0.0 0.0 {0.0 0.0 0.020375013} 10.837841 {0.0 0.0 0.0} -0.0016691721 0.09998653 0.0;
frame 1.284; moveto -1.0 { -1000 29 12 87.14} 100.0 0.0 0.0 {0.0 0.0 0.020375013} 10.837841 {0.0 0.0 0.0} -0.0016691721 0.09998653 0.0;
frame 1.285; moveto -1.0 { -1000 29 12 87.14} 100.0 0.0 0.0 {0.0 0.0 0.020375013} 10.837841 {0.0 0.0 0.0} -0.0016691721 0.09998653 0.0;
frame 1.286; moveto -1.0 { -1000 29 12 87.14} 100.0 0.0 0.0 {0.0 0.0 0.020375013} 10.837841 {0.0 0.0 0.0} -0.0016691721 0.09998653 0.0;
frame 1.287; moveto -1.0 { -1000 29 12 87.14} 100.0 0.0 0.0 {0.0 0.0 0.020375013} 10.837841 {0.0 0.0 0.0} -0.0016691721 0.09998653 0.0;
frame 1.288; moveto -1.0 { -1000 29 12 87.14} 100.0 0.0 0.0 {0.0 0.0 0.020375013} 10.837841 {0.0 0.0 0.0} -0.0016691721 0.09998653 0.0;
frame 1.289; moveto -1.0 { -1000 29 12 87.14} 100.0 0.0 0.0 {0.0 0.0 0.020375013} 10.837841 {0.0 0.0 0.0} -0.0016691721 0.09998653 0.0;
frame 1.290; moveto -1.0 { -1000 29 12 87.14} 100.0 0.0 0.0 {0.0 0.0 0.020375013} 10.837841 {0.0 0.0 0.0} -0.0016691721 0.09998653 0.0;
frame 1.291; moveto -1.0 { -1000 29 12 87.14} 100.0 0.0 0.0 {0.0 0.0 0.020375013} 10.837841 {0.0 0.0 0.0} -0.0016691721 0.09998653 0.0;
frame 1.292; moveto -1.0 { -1000 29 12 87.14} 100.0 0.0 0.0 {0.0 0.0 0.020375013} 10.837841 {0.0 0.0 0.0} -0.0016691721 0.09998653 0.0;
frame 1.293; moveto -1.0 { -1000 29 12 87.14} 100.0 0.0 0.0 {0.0 0.0 0.020375013} 10.837841 {0.0 0.0 0.0} -0.0016691721 0.09998653 0.0;
frame 1.294; moveto -1.0 { -1000 29 12 87.14} 100.0 0.0 0.0 {0.0 0.0 0.020375013} 10.837841 {0.0 0.0 0.0} -0.0016691721 0.09998653 0.0;
set fontScaling false;
}
function _setFrameState() {
- frame state;
- modelCount 310;
- first 1.1;
- last 1.310;
frame RANGE 1.1 1.310; animation DIRECTION +1; animation FPS 10; animation MODE ONCE 0.0 0.0; frame 1.281; animation OFF;
}
function _setPerspectiveState() {
set perspectiveModel 11;
set scaleAngstromsPerInch 0.0;
set perspectiveDepth true;
set visualRange 5.0;
set cameraDepth 3.0;
boundbox corners {-6.4248195 -6.424252 -6.9993925} {6.4248195 6.424252 7.040143} # volume = 2317.9082;
center {0.0 0.0 0.020375013};
moveto 0.0 { -277 673 685 148.94} 100.0 0.0 0.0 {0.0 0.0 0.020375013} 10.837841 {0.0 0.0 0.0} 4.531163E-4 0.10011922 0.0;;
slab 100;depth 0;
set spinX 0; set spinY 30; set spinZ 0; set spinFps 30; set navX 0; set navY 0; set navZ 0; set navFps 10;
vibration ON;
}
function _setSelectionState() {
select ({0:31929});
set hideNotSelected false;
}
function _setState() {
initialize; set refreshing false; _setWindowState; _setFileState; _setVariableState; _setModelState; _setFrameState; _setPerspectiveState; _setSelectionState; set refreshing true; set antialiasDisplay false; set antialiasTranslucent true; set antialiasImages true;
}
_setState;
</script> <uploadedFileContents>PBE-DZP_Niporph_Nitropyridin_2zu1_coplanar-t.IR.pseudo.log</uploadedFileContents> </jmolApplet>
</jmol> Nitropy asymm Streckschwingung des NO2 bei 1579 Wellenzahlen
$vibrational spectrum
# mode symmetry wave number IR intensity selection rules
# cm**(-1) km/mol IR RAMAN
1 0.00 0.00000 - -
2 0.00 0.00000 - -
3 0.00 0.00000 - -
4 0.00 0.00000 - -
5 0.00 0.00000 - -
6 0.00 0.00000 - -
7 a 9.02 0.00421 YES YES
8 b 11.23 0.00137 YES YES
9 b 12.38 0.19681 YES YES
10 a 16.08 0.02680 YES YES
11 b 18.24 0.00295 YES YES
12 a 19.98 0.00205 YES YES
13 b 22.15 0.00333 YES YES
14 a 22.83 0.00001 YES YES
15 b 23.29 0.00512 YES YES
16 a 24.13 0.00021 YES YES
17 b 25.34 0.00067 YES YES
18 b 26.76 0.06526 YES YES
19 b 31.23 0.17976 YES YES
20 a 35.25 0.00037 YES YES
21 a 37.06 0.00115 YES YES
22 b 37.58 0.01887 YES YES
23 a 47.60 0.00525 YES YES
24 b 51.67 0.01605 YES YES
25 a 58.32 0.00027 YES YES
26 a 60.25 0.00753 YES YES
27 a 72.46 0.02347 YES YES
28 a 74.28 0.00299 YES YES
29 a 75.83 0.00252 YES YES
30 b 76.34 0.00139 YES YES
31 a 80.58 0.00002 YES YES
32 b 89.36 3.34195 YES YES
33 a 92.69 0.00038 YES YES
34 b 102.31 0.03904 YES YES
35 b 103.38 0.07366 YES YES
36 b 106.81 0.53056 YES YES
37 a 106.99 0.00025 YES YES
38 b 108.34 0.19534 YES YES
39 b 113.41 0.03678 YES YES
40 b 117.65 4.15475 YES YES
41 a 125.33 0.00319 YES YES
42 a 127.44 0.39998 YES YES
43 b 130.51 0.10334 YES YES
44 a 130.55 0.00031 YES YES
45 b 131.01 0.06235 YES YES
46 a 131.02 0.00006 YES YES
47 a 139.85 0.06468 YES YES
48 a 148.42 0.93461 YES YES
49 b 162.66 0.17894 YES YES
50 b 165.51 0.24259 YES YES
51 a 169.97 0.00022 YES YES
52 b 173.92 0.02271 YES YES
53 a 180.20 0.00010 YES YES
54 b 182.14 0.13523 YES YES
55 b 188.58 0.16874 YES YES
56 b 212.87 4.58724 YES YES
57 a 213.47 0.00358 YES YES
58 b 222.22 1.33599 YES YES
59 b 233.74 0.40820 YES YES
60 b 236.55 2.77117 YES YES
61 b 242.39 0.29912 YES YES
62 a 242.86 0.00153 YES YES
63 a 246.99 21.62740 YES YES
64 b 254.06 0.06326 YES YES
65 a 256.30 1.18967 YES YES
66 b 259.68 0.02715 YES YES
67 a 261.09 0.43224 YES YES
68 a 262.16 0.00079 YES YES
69 b 263.46 0.00038 YES YES
70 b 263.49 0.00314 YES YES
71 a 263.78 1.02473 YES YES
72 a 264.74 0.05370 YES YES
73 a 267.19 0.00562 YES YES
74 b 267.44 0.01152 YES YES
75 b 267.53 0.00528 YES YES
76 a 272.71 1.04415 YES YES
77 b 276.65 0.39476 YES YES
78 b 283.07 0.08224 YES YES
79 b 286.24 0.01383 YES YES
80 a 287.87 0.01012 YES YES
81 b 289.97 0.09706 YES YES
82 a 301.70 2.08847 YES YES
83 b 303.32 0.26276 YES YES
84 b 303.56 0.11736 YES YES
85 a 303.82 0.03566 YES YES
86 a 311.36 0.17546 YES YES
87 a 337.15 0.00000 YES YES
88 b 337.45 3.03227 YES YES
89 b 338.61 3.84442 YES YES
90 b 369.14 2.56834 YES YES
91 a 370.49 0.00337 YES YES
92 b 371.15 2.38956 YES YES
93 a 382.67 0.00014 YES YES
94 a 387.79 0.16045 YES YES
95 a 388.98 0.41622 YES YES
96 a 390.15 1.30988 YES YES
97 b 392.07 0.69850 YES YES
98 a 392.40 0.00031 YES YES
99 b 392.56 0.57338 YES YES
100 a 392.81 0.02464 YES YES
101 a 393.81 0.01853 YES YES
102 a 399.77 0.01566 YES YES
103 a 407.07 0.00072 YES YES
104 b 415.82 4.26121 YES YES
105 b 416.26 4.70408 YES YES
106 a 429.60 0.86420 YES YES
107 b 430.05 0.01167 YES YES
108 b 430.20 0.03362 YES YES
109 a 430.88 0.00455 YES YES
110 a 433.68 0.23368 YES YES
111 b 444.81 5.13959 YES YES
112 b 446.27 4.32921 YES YES
113 b 465.92 0.01588 YES YES
114 b 469.28 1.20762 YES YES
115 a 479.81 0.00015 YES YES
116 a 487.64 0.07419 YES YES
117 b 499.17 0.90909 YES YES
118 a 499.48 0.07600 YES YES
119 b 499.57 0.93856 YES YES
120 b 516.33 0.14621 YES YES
121 b 517.17 0.66472 YES YES
122 a 524.51 3.32556 YES YES
123 b 525.11 0.15068 YES YES
124 b 527.52 0.23837 YES YES
125 a 528.44 0.07649 YES YES
126 a 569.96 0.00453 YES YES
127 b 570.39 0.06829 YES YES
128 b 571.62 0.42651 YES YES
129 a 571.76 0.00238 YES YES
130 a 634.61 0.00121 YES YES
131 b 640.72 1.07753 YES YES
132 b 641.06 0.86613 YES YES
133 a 645.70 0.00407 YES YES
134 b 645.83 0.00595 YES YES
135 a 648.02 0.04254 YES YES
136 b 648.97 0.25600 YES YES
137 a 649.13 0.00000 YES YES
138 b 649.21 0.02466 YES YES
139 b 649.84 0.25689 YES YES
140 a 660.22 0.00098 YES YES
141 b 667.87 0.59228 YES YES
142 b 668.51 0.04994 YES YES
143 a 682.05 0.06424 YES YES
144 a 684.75 23.31640 YES YES
145 a 685.89 2.25873 YES YES
146 b 689.10 7.90681 YES YES
147 b 689.64 16.32140 YES YES
148 a 693.59 0.06051 YES YES
149 b 695.97 3.71226 YES YES
150 b 701.63 7.62123 YES YES
151 a 705.72 13.64767 YES YES
152 a 713.48 4.29877 YES YES
153 b 715.48 0.01923 YES YES
154 b 716.79 1.01336 YES YES
155 a 722.21 0.01131 YES YES
156 a 725.99 0.00060 YES YES
157 b 740.05 1.87868 YES YES
158 b 741.41 15.38599 YES YES
159 b 752.28 141.57929 YES YES
160 b 753.57 136.46292 YES YES
161 a 760.62 24.71369 YES YES
162 b 766.47 7.33117 YES YES
163 b 767.78 0.19319 YES YES
164 a 771.61 0.63974 YES YES
165 a 778.95 0.01872 YES YES
166 a 790.26 0.29620 YES YES
167 b 795.85 0.17385 YES YES
168 b 798.46 0.42371 YES YES
169 a 802.30 14.45436 YES YES
170 a 812.67 0.45097 YES YES
171 b 823.43 1.44443 YES YES
172 b 824.31 0.73172 YES YES
173 a 836.86 0.13068 YES YES
174 a 848.52 95.28003 YES YES
175 a 849.15 7.59225 YES YES
176 b 853.42 2.43143 YES YES
177 b 855.34 8.57633 YES YES
178 a 857.07 0.07555 YES YES
179 a 858.31 0.25989 YES YES
180 b 894.41 0.30803 YES YES
181 a 895.36 0.00019 YES YES
182 b 898.25 0.02382 YES YES
183 a 899.44 0.00026 YES YES
184 a 912.00 1.98767 YES YES
185 b 929.27 213.70004 YES YES
186 b 929.68 212.70349 YES YES
187 a 953.63 0.01099 YES YES
188 b 965.80 0.63053 YES YES
189 b 966.70 1.88920 YES YES
190 a 973.24 0.19360 YES YES
191 a 973.94 0.32454 YES YES
192 a 991.72 60.28631 YES YES
193 b 994.19 9.64961 YES YES
194 a 994.53 44.15504 YES YES
195 a 995.71 24.06699 YES YES
196 a 996.20 2.60983 YES YES
197 b 997.63 6.68147 YES YES
198 a 998.95 440.16612 YES YES
199 b 1003.58 29.65786 YES YES
200 b 1005.48 25.40696 YES YES
201 a 1007.97 4.43225 YES YES
202 a 1027.56 20.83584 YES YES
203 a 1030.19 17.39513 YES YES
204 a 1036.44 0.28639 YES YES
205 b 1041.73 17.60866 YES YES
206 b 1043.75 10.09635 YES YES
207 a 1048.43 0.00837 YES YES
208 a 1051.76 0.01654 YES YES
209 b 1054.98 219.24097 YES YES
210 b 1056.83 185.26089 YES YES
211 b 1061.56 0.56692 YES YES
212 b 1062.81 2.96962 YES YES
213 a 1071.81 3.14686 YES YES
214 a 1101.42 71.38775 YES YES
215 a 1102.21 6.34241 YES YES
216 a 1145.37 0.65667 YES YES
217 a 1161.68 9.33201 YES YES
218 b 1162.04 4.42736 YES YES
219 b 1165.56 6.23990 YES YES
220 a 1166.02 7.76879 YES YES
221 b 1168.38 29.16157 YES YES
222 b 1169.93 22.92181 YES YES
223 a 1171.51 0.00923 YES YES
224 a 1186.76 3.06342 YES YES
225 a 1190.35 1.93691 YES YES
226 b 1219.68 25.88663 YES YES
227 b 1219.82 24.57157 YES YES
228 a 1219.95 0.86826 YES YES
229 a 1251.31 1.33699 YES YES
230 b 1279.12 0.57723 YES YES
231 b 1280.91 1.72379 YES YES
232 b 1301.26 0.30629 YES YES
233 b 1305.26 1.71076 YES YES
234 a 1312.26 0.11230 YES YES
235 a 1314.72 0.11034 YES YES
236 b 1319.57 13.67285 YES YES
237 b 1320.84 7.52668 YES YES
238 a 1322.38 0.00928 YES YES
239 b 1325.02 3.35584 YES YES
240 a 1325.74 0.15919 YES YES
241 b 1327.11 9.64416 YES YES
242 a 1331.98 645.50994 YES YES
243 a 1333.71 2.28869 YES YES
244 b 1342.13 10.16776 YES YES
245 b 1343.19 10.90017 YES YES
246 a 1343.45 0.39798 YES YES
247 a 1344.09 0.01326 YES YES
248 b 1349.02 67.25587 YES YES
249 b 1351.11 59.39731 YES YES
250 a 1353.33 0.00699 YES YES
251 a 1356.22 0.99358 YES YES
252 b 1392.35 3.14602 YES YES
253 b 1393.36 20.75685 YES YES
254 b 1424.60 28.12216 YES YES
255 b 1428.00 24.91008 YES YES
256 a 1429.86 0.51481 YES YES
257 a 1434.50 0.92120 YES YES
258 b 1453.29 56.26257 YES YES
259 b 1454.16 66.10338 YES YES
260 a 1456.29 6.20468 YES YES
261 a 1457.80 4.70384 YES YES
262 a 1468.62 0.13256 YES YES
263 a 1473.04 0.49600 YES YES
264 b 1500.53 60.90326 YES YES
265 a 1500.80 365.80924 YES YES
266 b 1502.02 70.84351 YES YES
267 a 1502.27 682.46114 YES YES
268 a 1512.56 0.22932 YES YES
269 b 1517.09 696.72341 YES YES
270 b 1517.49 630.80436 YES YES
271 a 1518.50 17.55629 YES YES
272 a 1520.24 0.18471 YES YES
273 a 1523.91 93.20565 YES YES
274 b 1528.49 33.86448 YES YES
275 b 1529.89 15.09165 YES YES
276 b 1541.26 59.59274 YES YES
277 b 1541.84 54.86659 YES YES
278 a 1560.81 0.00055 YES YES
279 a 1568.23 0.22025 YES YES
280 b 1579.31 32.55262 YES YES
281 b 1579.85 154.17121 YES YES
282 a 1588.77 2.41558 YES YES
283 a 1589.54 0.20599 YES YES
284 b 1619.45 1.02943 YES YES
285 a 1619.57 15.04724 YES YES
286 b 1620.09 1.00603 YES YES
287 a 1620.22 5.80031 YES YES
288 b 1621.75 22.05278 YES YES
289 b 1622.17 75.90755 YES YES
290 a 1636.91 0.50723 YES YES
291 b 1636.92 35.70114 YES YES
292 b 1637.41 31.13187 YES YES
293 a 1637.49 0.13190 YES YES
294 b 3139.54 1.12345 YES YES
295 a 3140.88 16.56893 YES YES
296 b 3152.94 1.10426 YES YES
297 a 3153.58 26.19790 YES YES
298 a 3166.80 3.84064 YES YES
299 b 3167.52 3.45497 YES YES
300 a 3167.53 0.20569 YES YES
301 b 3167.56 5.11327 YES YES
302 a 3172.84 0.03828 YES YES
303 b 3172.88 3.61690 YES YES
304 a 3174.50 0.01135 YES YES
305 b 3174.52 3.25487 YES YES
306 b 3191.03 9.14819 YES YES
307 a 3191.03 0.00078 YES YES
308 b 3192.63 9.07271 YES YES
309 a 3192.67 0.00114 YES YES